Dong Gao, Min Zheng, Genmei Lin, Wenyu Fang, Jing Huang, Jianguo Lu*, Xiaowen Sun. Construction of high-density genetic map and mapping of sex determination loci in the yellow catfish (Pelteobagrus fulyidraco)
Shizhu Li, Genmei Lin, Wenyu Fang, Peilin Huang, Dong Gao, Jing Huang, Jingui Xie, Jianguo Lu*. Gonadal transcriptome analysis of sex-related genes in the protandrous yellowfin seabream (Acanthopagrus latus). Frontiers in Genetics, 2020, 11.
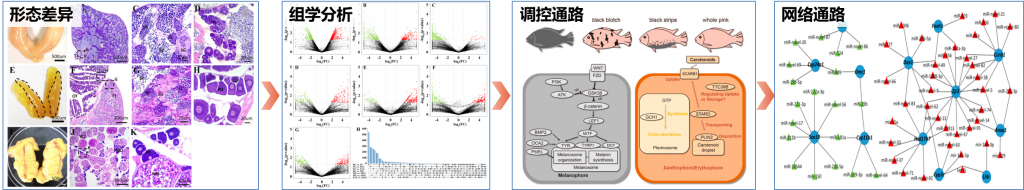
Wenyu Fang, Junrou Huang, Shizhu Li, Jianguo Lu*. Identification of pigment genes (melanin, carotenoid and pteridine) associated with skin color variant in red tilapia using transcriptome analysis. Aquaculture, 2021, 547: 737429.
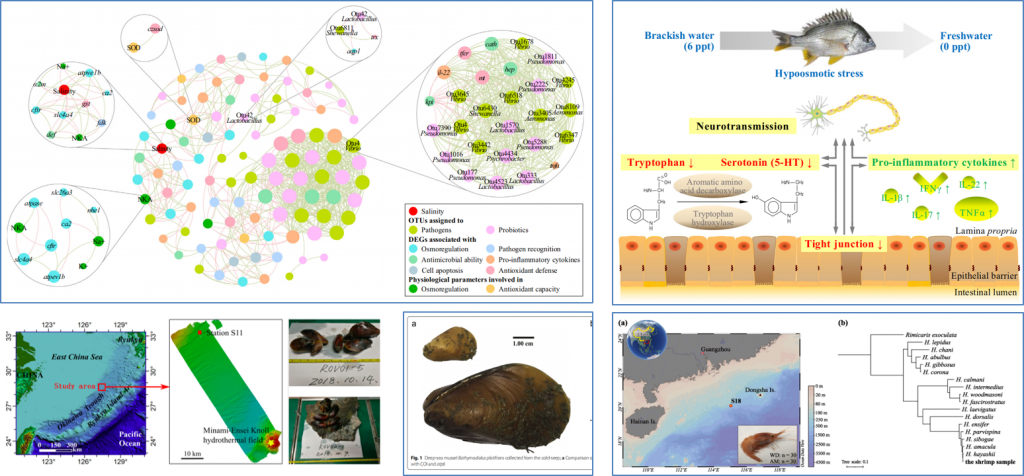
Genmei Lin, Shizhu Li, Junrou Huang, Dong Gao, Jianguo Lu*. Hypoosmotic stress induced functional alternations of intestinal barrier integrity, inflammatory reactions, and neurotransmission along gut-brain axis in the yellowfin seabream (Acanthopagrus latus). Fish Physiology and Biochemistry, 2021, 47: 1725-1738.
Genmei Lin, Min Zheng, Dong Gao, Shizhu Li, Wenyu Fang, Jing Huang, Jingui Xie, Jingxiong Liu, Yijing Liu, Zhaohong Li, Jianguo Lu*. Hypoosmotic stress induced tissue-specific immune responses of yellowfin seabream (Acanthopagrus latus) revealed by transcriptomic analysis. Fish & Shellfish Immunology, 2020, 99: 473-482.
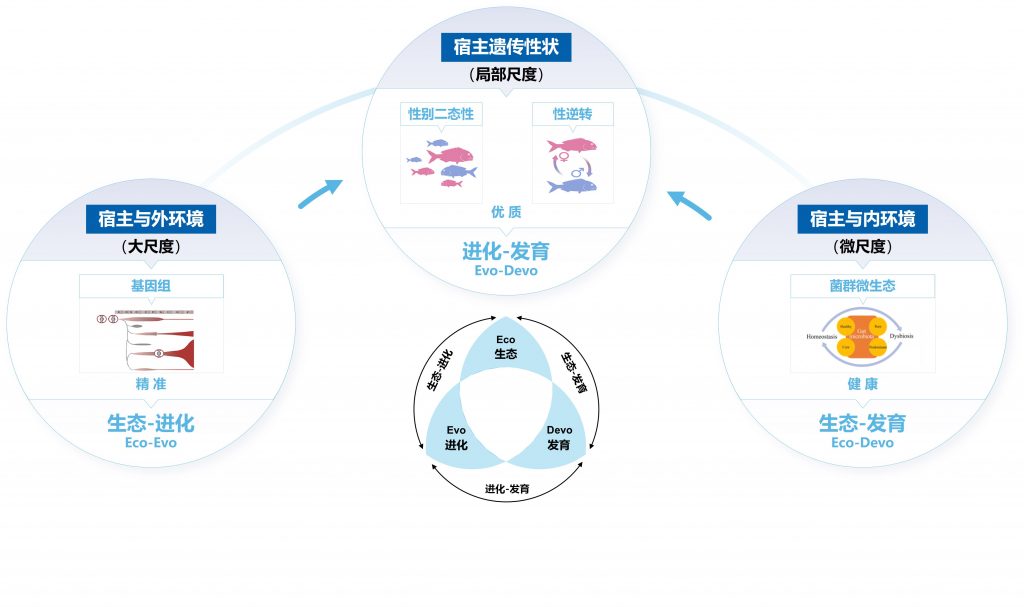
Genmei Lin, Dong Gao, Jianguo Lu*, Xiaowen Sun. Transcriptome profiling insights the sexual dimorphism of gene expression patterns during gonad differentiation in the half-smooth tongue sole (Cynoglossus semilaevis). Marine Biotechnology, 2021, 23: 18-30.
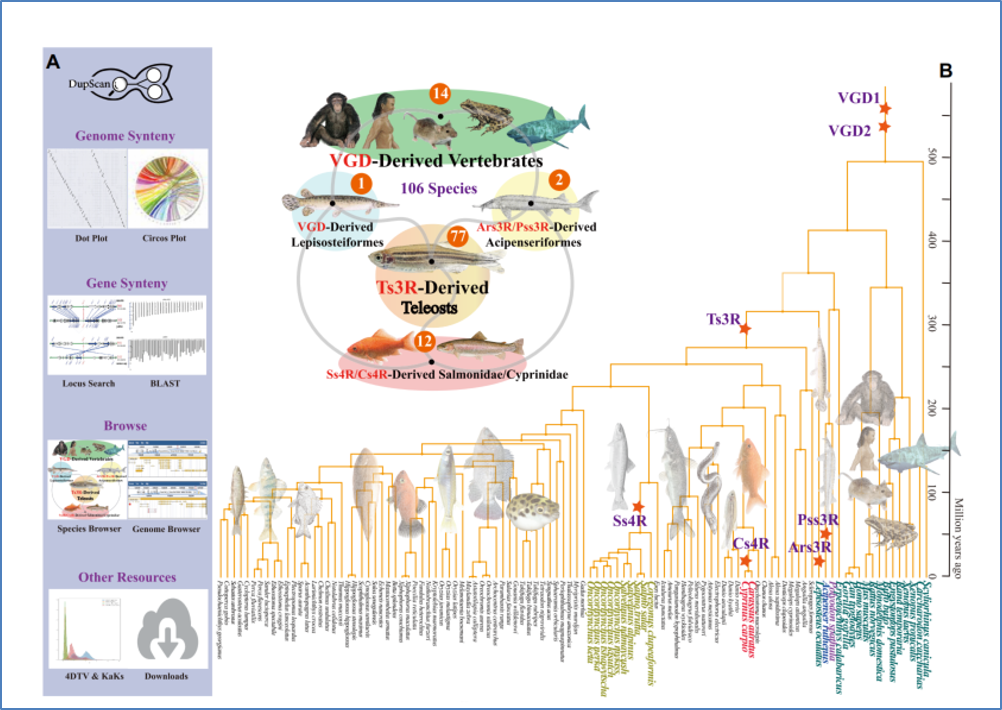
Jianguo Lu, Wenyu Fang, Junrou Huang, Shizhu Li. The application of genome editing technology in fish. Marine Life Science & Technology, 2021, 3: 326-346.
李石竹, 方文宇, 骆明飞, 卢建国*. 黄颡鱼GnRHR基因的克隆和表达及CRISPR/Cas9构建GnRHR基因敲除突变体. 大连海洋大学学报, 2021, 36(3): 383-392.
Gao D, Huang J, Lin G, Lu J*. A time-course transcriptome analysis of gonads from yellow catfish (Pelteobagrus fulvidraco) reveals genes associated with gonad development[J]. BMC Genomics, 2022, 23(S1).
Jianguo Lu, Dong Gao, Ying Sims, Wenyu Fang, Joanna Collins, James Torrance, Genmei Lin, et al. Chromosome-level Genome Assembly of Acanthopagrus latus Provides Insights into Salinity Stress Adaptation of Sparidae. Mar Biotechnol, 2022,24: 655-660.
Fang W, Huang J, Li S, Lu J*. Identification of pigment genes (melanin, carotenoid and pteridine) associated with skin color variant in red tilapia using transcriptome analysis[J]. Aquaculture, 2022, 547: 737429.
Marco Podobnik, Ajeet P. Singh, Zhenqiang Fu, Christopher M. Dooley, Hans Georg Frohnhöfer, Magdalena Firlej, Sarah J. Stednitz, Hadeer Elhabashy, Simone Weyand, John R. Weir, Jianguo Lu, Christiane Nüsslein-Volhard, Uwe Irion. kcnj13 regulates pigment cell shapes in zebrafish and has diverged by cis-regulatory evolution between Danio species[J]. Development, 2023, 150(16).