我们是中山大学海洋学院海洋生态基因组学团队。 海洋生态基因组学团队利用基因组学方法研究海洋生态问题,阐明海洋生物适应极端环境(冷渗、热液喷口、深海区域)的机制,并通过基因数据分析,探索海洋生物性别决定、抗病、生长、色素沉着等重要经济性状的相关功能,从而与产业相结合。 我们的工作包括:

Marine Ecological Genomics Lab
中山大学海洋科学学院 海洋生态基因组实验室

实验室简介
(1)海洋鱼类对不同生境的适应机制、进化模式和性别调节机制; (2)功能基因对极端环境(冷泉和热液区)生物适应的调控机制。 (3)海洋微生物对富水合物海区甲烷气体运移机制的影响; (4)深海、超深海、深渊海域海洋生态适应机制; (5)滨海湿地红树林生态系统中海洋生物的适应机制。
代表性论文
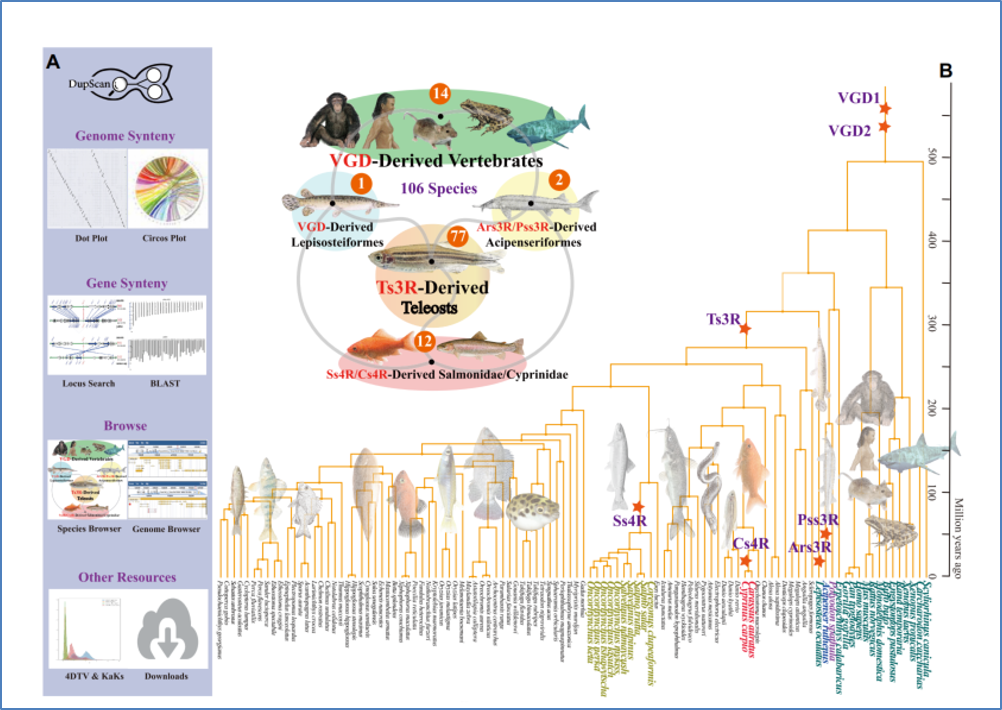
- Lu J*, et al. DupScan: predicting and visualizing vertebrate genome duplication database. Nucleic Acids Res. 2022. 1区Top, IF=19.16
- Fang W … Lu J*. Identification of pigment genes (melanin, carotenoid and pteridine) associated with skin color variant in red tilapia using transcriptome analysis. Aquaculture. 2022. 1区Top
- Genmei Lin … Jianguo Lu*. Response of gut microbiota and immune function to hypoosmotic stress in the yellowfin seabream (Acanthopagrus latus). Science of the Total Environment, 2020. 1区Top
- Genmei Lin, Xianbiao Lin*. Bait input altered microbial community structure and increased greenhouse gases production in coastal wetland sediment. Water Research, 2022. 1区Top
- Lu, J*.et al. Chromosome-level Genome Assembly of Acanthopagrus latus Provides Insights into Salinity Stress Adaptation of Sparidae. Mar Biotechnol 24, 655–660 (2022).
- Lin G … Lu J*. Characterization of tissue-associated bacterial community of two Bathymodiolus species from the adjacent cold seep and hydrothermal vent environments. Science of the Total Environment. 2021
(1)构建了规模最大的脊椎动物多组学融合平台,为功能基因挖掘提供精准数据
Lu J*, et al. DupScan: predicting and visualizing vertebrate genome duplication database. Nucleic Acids Res. 2022. 1区Top, IF=19.16
整合106个高质量脊椎动物多组学数据,搭建了一站式多组学分析平台DupScan数据库 技术性突破: 1. 多组学融合技术 2. 性状功能基因挖掘与应用
主要优势 专为鱼类多倍体化分析设计 收录数据较全面 可视化、可交互、可相互配合 界面友 好配套视频与文字教程
(2)阐明了海洋鱼类重要经济性状功能基因的遗传机制,为品种改良和种质创新提供依据
技术性突破:
以黄颡鱼、黄鳍鲷、红罗非鱼、半滑舌鳎重要经济水产物种为研究对象,构建挖掘性别二态性、性逆转鱼类相关的功能基因核心技术。
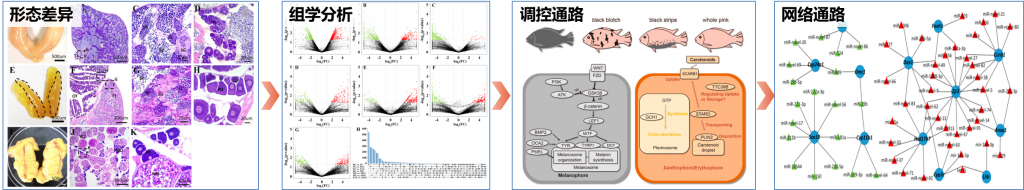
Dong Gao, Min Zheng, Genmei Lin, Wenyu Fang, Jing Huang, Jianguo Lu*, Xiaowen Sun. Construction of high-density genetic map and mapping of sex determination loci in the yellow catfish (Pelteobagrus fulyidraco)
Shizhu Li, Genmei Lin, Wenyu Fang, Peilin Huang, Dong Gao, Jing Huang, Jingui Xie, Jianguo Lu*. Gonadal transcriptome analysis of sex-related genes in the protandrous yellowfin seabream (Acanthopagrus latus). Frontiers in Genetics, 2020, 11.
Wenyu Fang, Junrou Huang, Shizhu Li, Jianguo Lu*. Identification of pigment genes (melanin, carotenoid and pteridine) associated with skin color variant in red tilapia using transcriptome analysis. Aquaculture, 2021, 547: 737429.
Genmei Lin, Shizhu Li, Junrou Huang, Dong Gao, Jianguo Lu*. Hypoosmotic stress induced functional alternations of intestinal barrier integrity, inflammatory reactions, and neurotransmission along gut-brain axis in the yellowfin seabream (Acanthopagrus latus). Fish Physiology and Biochemistry, 2021, 47: 1725-1738.
Genmei Lin, Min Zheng, Dong Gao, Shizhu Li, Wenyu Fang, Jing Huang, Jingui Xie, Jingxiong Liu, Yijing Liu, Zhaohong Li, Jianguo Lu*. Hypoosmotic stress induced tissue-specific immune responses of yellowfin seabream (Acanthopagrus latus) revealed by transcriptomic analysis. Fish & Shellfish Immunology, 2020, 99: 473-482.
Genmei Lin, Dong Gao, Jianguo Lu*, Xiaowen Sun. Transcriptome profiling insights the sexual dimorphism of gene expression patterns during gonad differentiation in the half-smooth tongue sole (Cynoglossus semilaevis). Marine Biotechnology, 2021, 23: 18-30.
Jianguo Lu, Wenyu Fang, Junrou Huang, Shizhu Li. The application of genome editing technology in fish. Marine Life Science & Technology, 2021, 3: 326-346.
李石竹, 方文宇, 骆明飞, 卢建国*. 黄颡鱼GnRHR基因的克隆和表达及CRISPR/Cas9构建GnRHR基因敲除突变体. 大连海洋大学学报, 2021, 36(3): 383-392.
Gao D, Huang J, Lin G, Lu J*. A time-course transcriptome analysis of gonads from yellow catfish (Pelteobagrus fulvidraco) reveals genes associated with gonad development[J]. BMC Genomics, 2022, 23(S1).
Jianguo Lu, Dong Gao, Ying Sims, Wenyu Fang, Joanna Collins, James Torrance, Genmei Lin, et al. Chromosome-level Genome Assembly of Acanthopagrus latus Provides Insights into Salinity Stress Adaptation of Sparidae. Mar Biotechnol, 2022,24: 655-660.
Fang W, Huang J, Li S, Lu J*. Identification of pigment genes (melanin, carotenoid and pteridine) associated with skin color variant in red tilapia using transcriptome analysis[J]. Aquaculture, 2022, 547: 737429.
Marco Podobnik, Ajeet P. Singh, Zhenqiang Fu, Christopher M. Dooley, Hans Georg Frohnhöfer, Magdalena Firlej, Sarah J. Stednitz, Hadeer Elhabashy, Simone Weyand, John R. Weir, Jianguo Lu, Christiane Nüsslein-Volhard, Uwe Irion. kcnj13 regulates pigment cell shapes in zebrafish and has diverged by cis-regulatory evolution between Danio species[J]. Development, 2023, 150(16).
(3)解析海洋渔业生物的环境适应性机制,为开发优质性状渔业新品种提供技术支撑
以深海特殊生境和传统养殖环境下的深海贻贝、深海虾、黄鳍鲷等为研究对象,进行多组学尺度下海洋渔业生物对胁迫环境的适应策略研究。
技术创新:宿主本身、体内肠道微生物、环境微生物,三者融合,探究适应性机制。
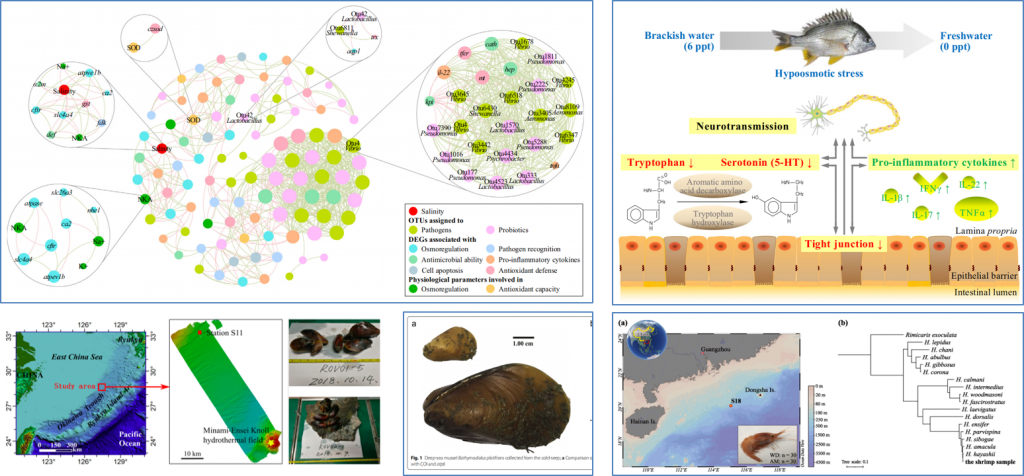
Genmei Lin, Min Zheng, Shizhu Li, Jingui Xie, Wenyu Fang, Dong Gao, Jing Huang, Jianguo Lu*. Response of gut microbiota and immune function to hypoosmotic stress in the yellowfin seabream (Acanthopagrus latus). Science of the Total Environment, 2020, 745: 140976.
Genmei Lin, Jianguo Lu*, Zhilei Sun, Jingui Xie, Junrou Huang, Ming Su, Nengyou Wu*. Characterization of tissue-associated bacterial community of two Bathymodiolus species from the adjacent cold seep and hydrothermal vent environments. Science of the Total Environment, 2021, 796: 149046.
Genmei Lin, Jing Huang, Jianguo Lu, Ming Su, Bangqi Hu, Xianbiao Lin*. Geochemical and microbial insights into vertical distributions of genetic potential of N-cycling processes in deep-sea sediments. Ecological Indicators, 2021, 125: 107461.
Huang, J., Huang, P., Lu J*. et al. Gene expression profiles provide insights into the survival strategies in deep-sea mussel (Bathymodiolus platifrons) of different developmental stages. BMC Genomics 23 (Suppl 1), 311 (2022).
Genmei Lin, Junrou Huang, Kunwen Luo, Xianbiao Lin, Ming Su, Jianguo Lu*. Bacterial, archaeal, and fungal community structure and interrelationships of deep-sea shrimp intestine and the surrounding sediment. Environmental Research, 2022, 205: 112461.
(4)环境微生物群落对共生微生物群落结构的影响及对能量代谢、细菌识别和抗菌活性等关键功能的调控机理
以代表性海洋生物为研究对象,通过培养实验和多组学测序分析手段,探究了环境微生物群落对海洋生物共生微生物群落结构的影响。

Genmei Lin, Yongni He, Jianguo Lu, Hui Chen, Jianxiang Feng*. Seasonal variations in soil physicochemical properties and microbial community structure influenced by Spartina alterniflora invasion and Kandelia obovata restoration. Science of the Total Environment, 2021, 797: 149213.
Hualin Liu, Vimalkumar Prajapati*, Shobha Prajapati, Harsh Bais, Jianguo Lu. Comparative genome analysis of Bacillus amyloliquefaciens focusing on phylogenomics, functional traits, and prevalence of antimicrobial and virulence genes. Frontiers in Genetics, 2021, 12: 724217.
Liu H, Lin G, Gao D, Lu J*. Geographic Scale Influences the Interactivities Between Determinism and Stochasticity in the Assembly of Sedimentary Microbial Communities on the South China Sea Shelf. Microb Ecol, 2022, 85(1): 121-136.
Genmei Lin, Xianbiao Lin*. Bait input altered microbial community structure and increased greenhouse gases production in coastal wetland sediment. Water Research, 2022, 218: 118520.
Genmei Lin, Jianguo Lu, Kunwen Luo, Yunxin Fang, Jiawei Liu, Xiang Ji, Shutong Ge, Jia Liu, Ming Su*. Characterization of bacterial and archaeal community structure in deep subsurface sediments in the Shenhu area, northern South China Sea. Marine and Petroleum Geology, 2022, 136: 105468.
Lin X, Lin G, Zheng Y, et al. Nitrogen mineralization and immobilization in surface sediments of coastal reclaimed aquaculture ecosystems[J]. Frontiers in Marine Science, 2023, 9.
Hualin Liu, Xueyu Cai, Kunwen Luo, Sihan Chen, Ming Su, Jianguo Lu. Microbial Diversity, Community Turnover, and Putative Functions in Submarine Canyon Sediments under the Action of Sedimentary Geology[J]. Microbiology Spectrum, 2023, 11(2).